Section: New Results
3D+t segmentation of single growing axons
Participants : Nadège Guiglielmoni, Caroline Medioni, Florence Besse, Xavier Descombes, Grégoire Malandain.
Our work is motivated by the study of developmental axonal remodeling, a genetically-controlled process characterized by a degeneration step followed by a rapid regrowth of axons. Here, we focus our interest on the axonal regrowth phase, which can be studied during brain development, using the fruit fly, Drosophila melanogaster, as a model system.
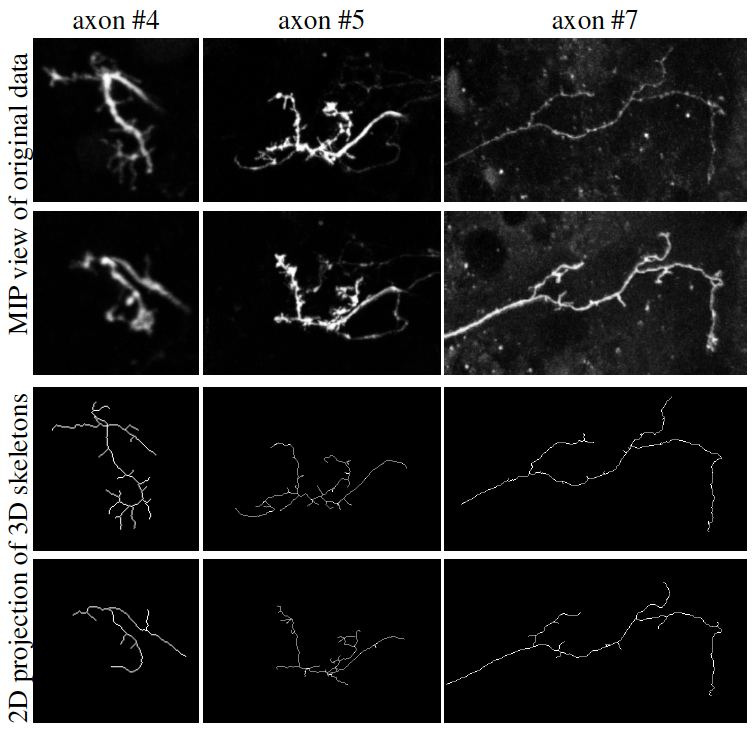
During the regrowth, small dynamical branches can be observed: they emanate from long stable branches and have generally a short lifetime. Such small branches may contribute to rebuild the axon connectivity during the adult stage. A better knowledge of the mechanisms controlling the dynamic of these branches may contribute to a better understanding of neuronal morphogenesis. In this work, we are particularly interested in the quantification of this process, for which the extraction of both the main and second branches is required.
Neuron tracing is still a challenge in neuroinformatics. Despite the huge progresses made during the last decades, this problem is still an open question. This is exacerbated with the development of new imaging techniques, that produce more and more images with improved quality and/or resolution. Among these, live-imaging techniques are more and more prominent. Indeed, acquisitions of 3D image sequences over long periods of time, in particular, have enabled neurobiologists to follow complex processes such as the development of neuronal populations. However, they produce time series of 3D volumes, for which there does not exist dedicated tracing approach.
Apart slight movements, the dynamic changes of axons are due to growing or retracting branches. Thus, we designed a topologically constrained tracking method that first ensures that the tree structure of the axon and its branches is preserved through the time sequence, and second enables a slight displacement of the axon (within an user-specified extend), while mimicing both the retraction and the growth of branches. Results are presented in figure 4.
|